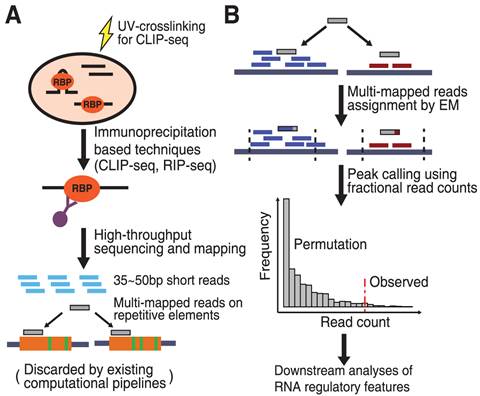
RNA structure, binding, and coordination in Arabidopsis - Foley - 2017 - WIREs RNA - Wiley Online Library

omniCLIP: probabilistic identification of protein-RNA interactions from CLIP -seq data | Genome Biology | Full Text
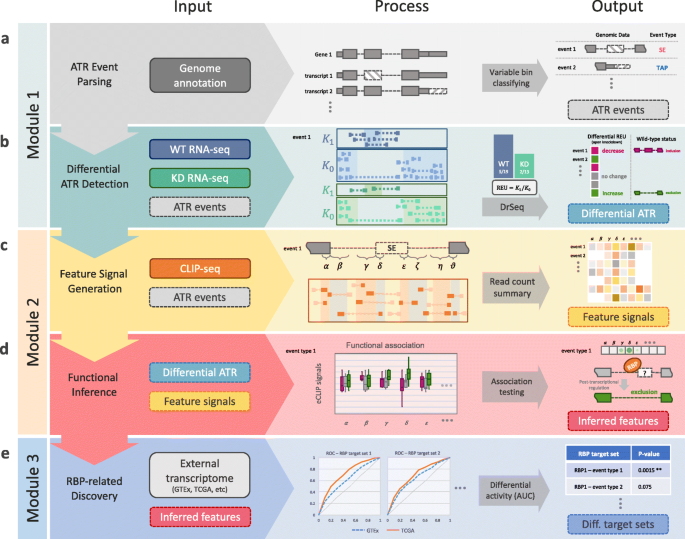
SURF: integrative analysis of a compendium of RNA-seq and CLIP-seq datasets highlights complex governing of alternative transcriptional regulation by RNA-binding proteins | Genome Biology | Full Text

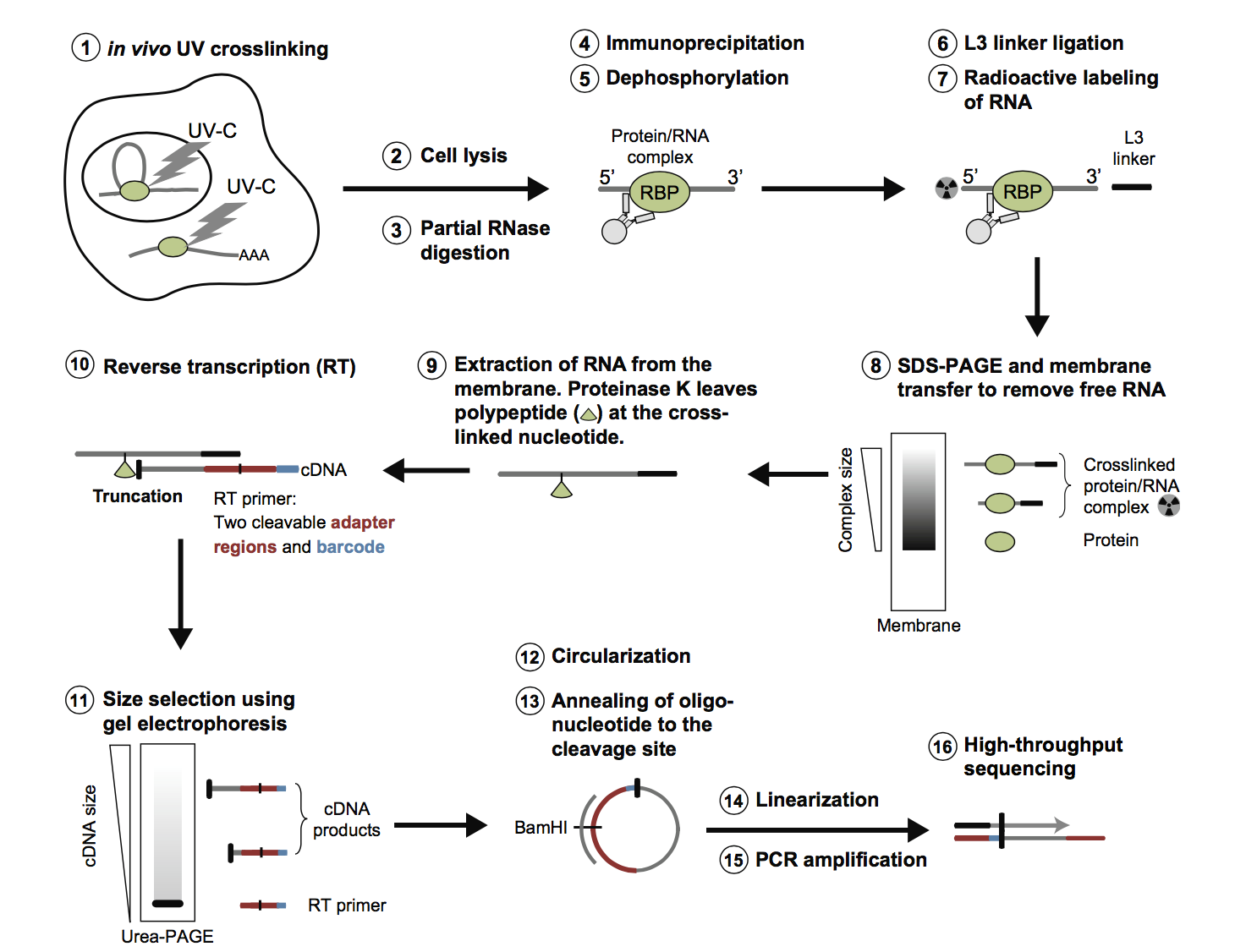
Main steps of the bioinformatics workflow to analyze CLIP-seq data with... | Download Scientific Diagram
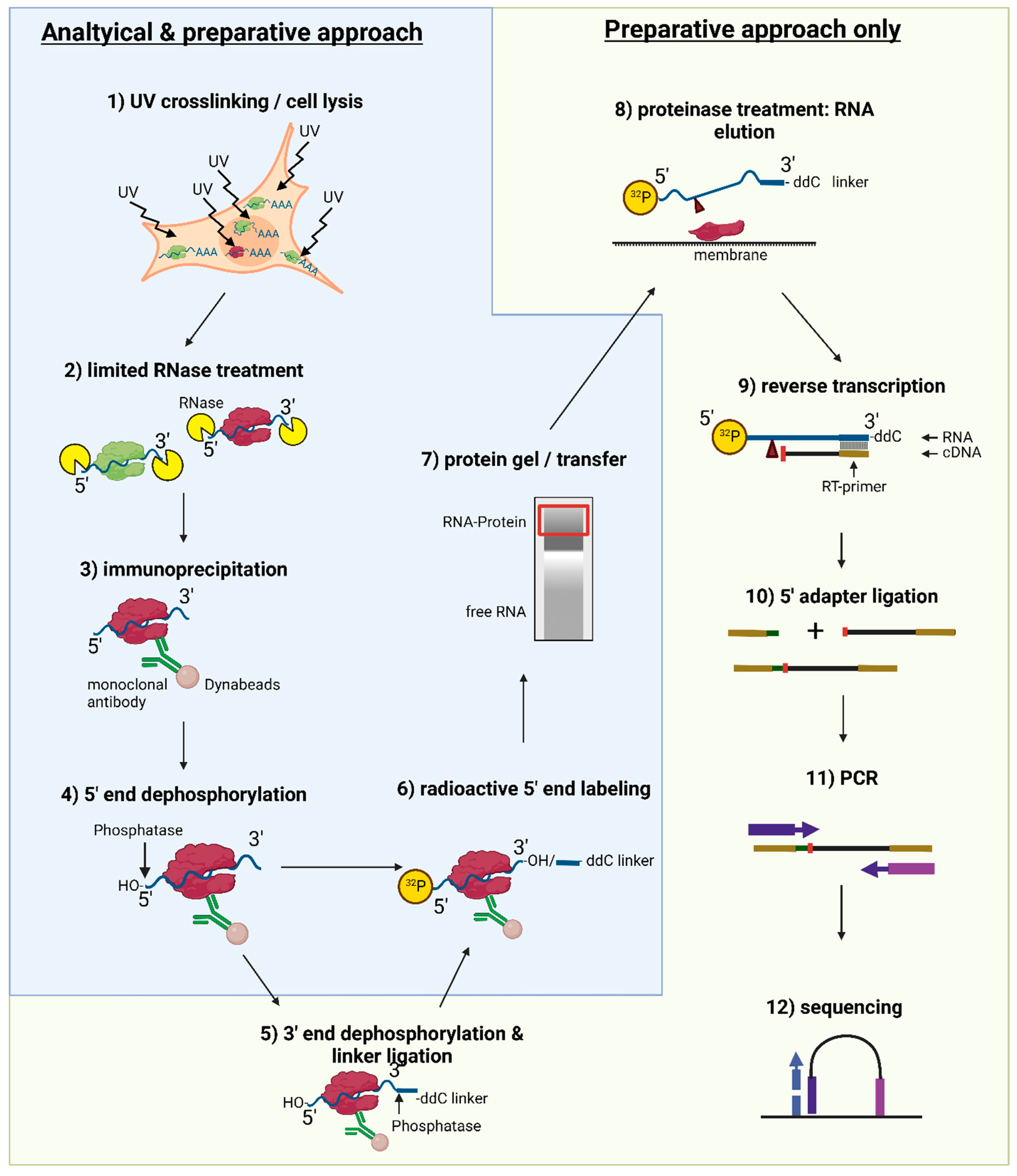
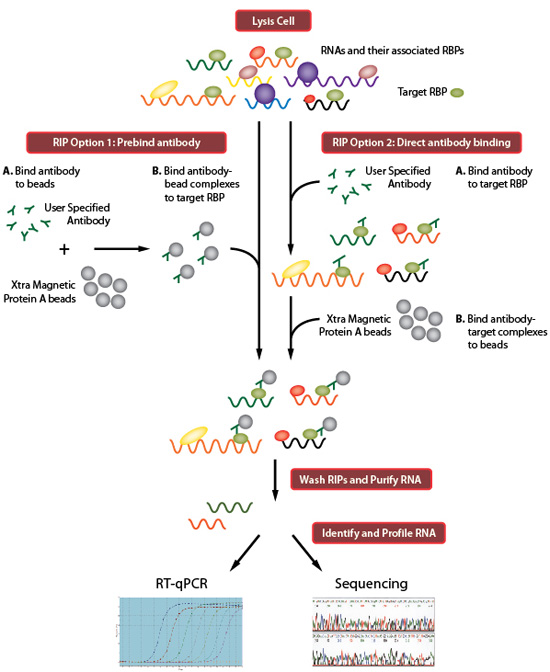
MPs | Free Full-Text | Studying miRNA–mRNA Interactions: An Optimized CLIP-Protocol for Endogenous Ago2-Protein


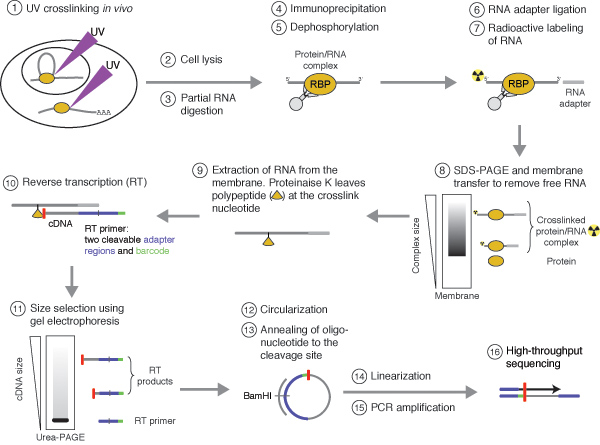
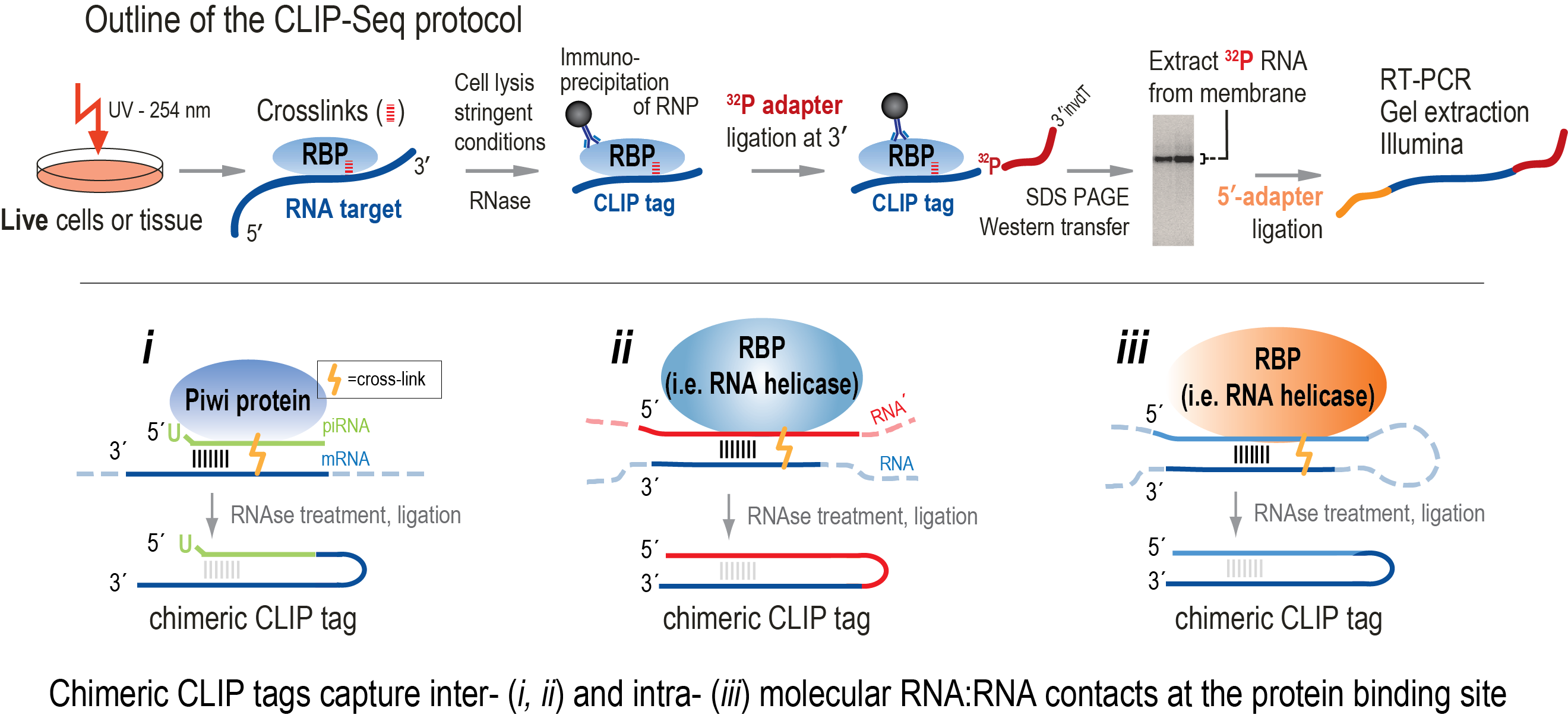
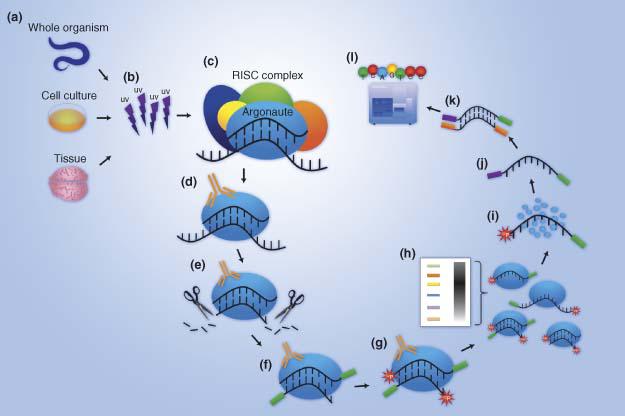

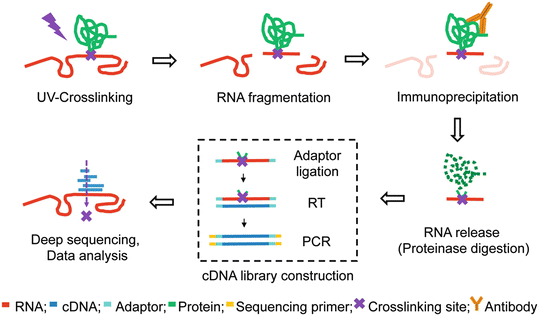


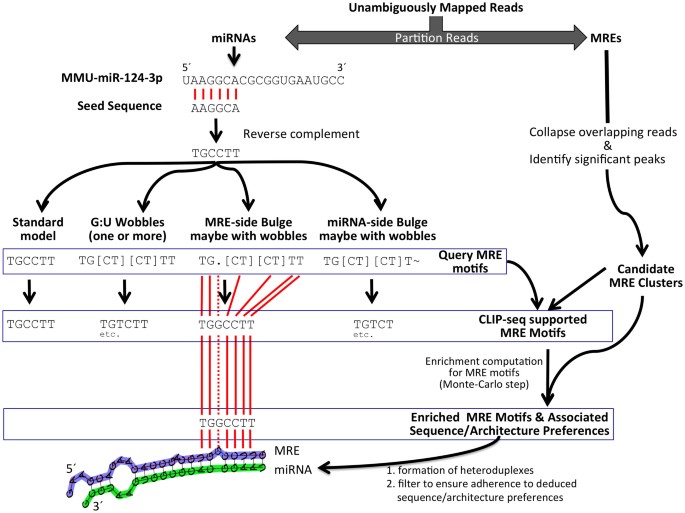
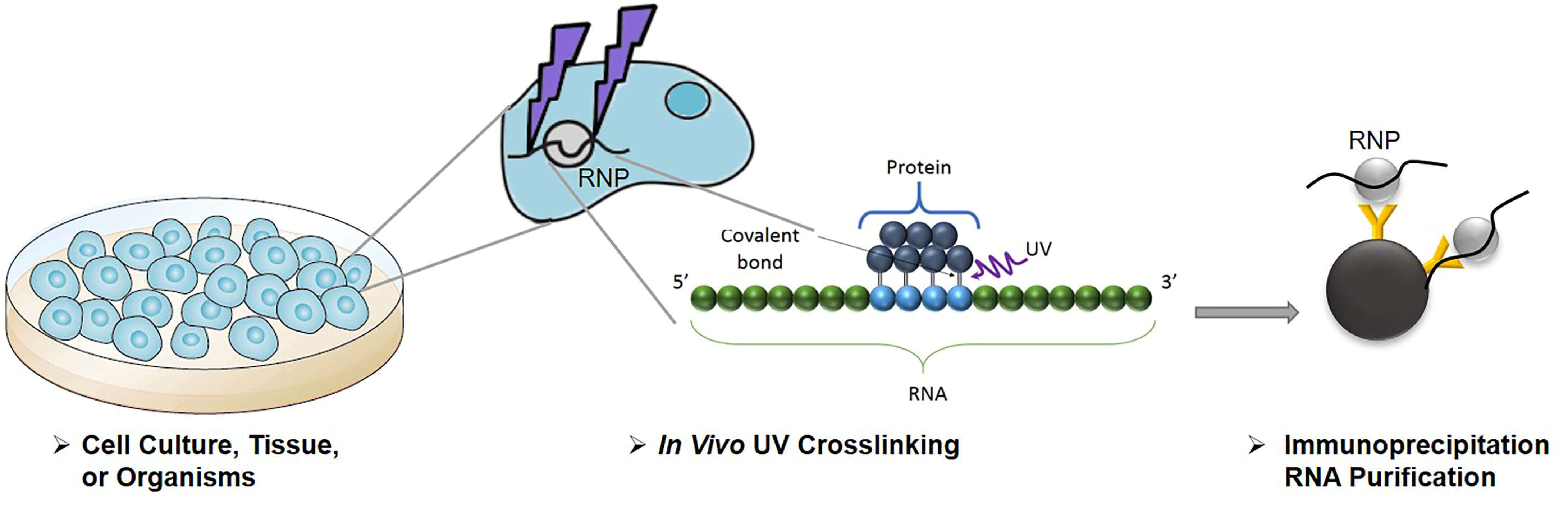
![PDF] Current methods in the analysis of CLIP-Seq data | Semantic Scholar PDF] Current methods in the analysis of CLIP-Seq data | Semantic Scholar](https://d3i71xaburhd42.cloudfront.net/e504d75c2c48b19613a796fb54083b71e3781289/2-Figure1-1.png)
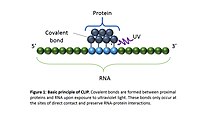
![PDF] Current methods in the analysis of CLIP-Seq data | Semantic Scholar PDF] Current methods in the analysis of CLIP-Seq data | Semantic Scholar](https://d3i71xaburhd42.cloudfront.net/e504d75c2c48b19613a796fb54083b71e3781289/3-Figure2-1.png)